Overview
What are Visual DNAs useful for?
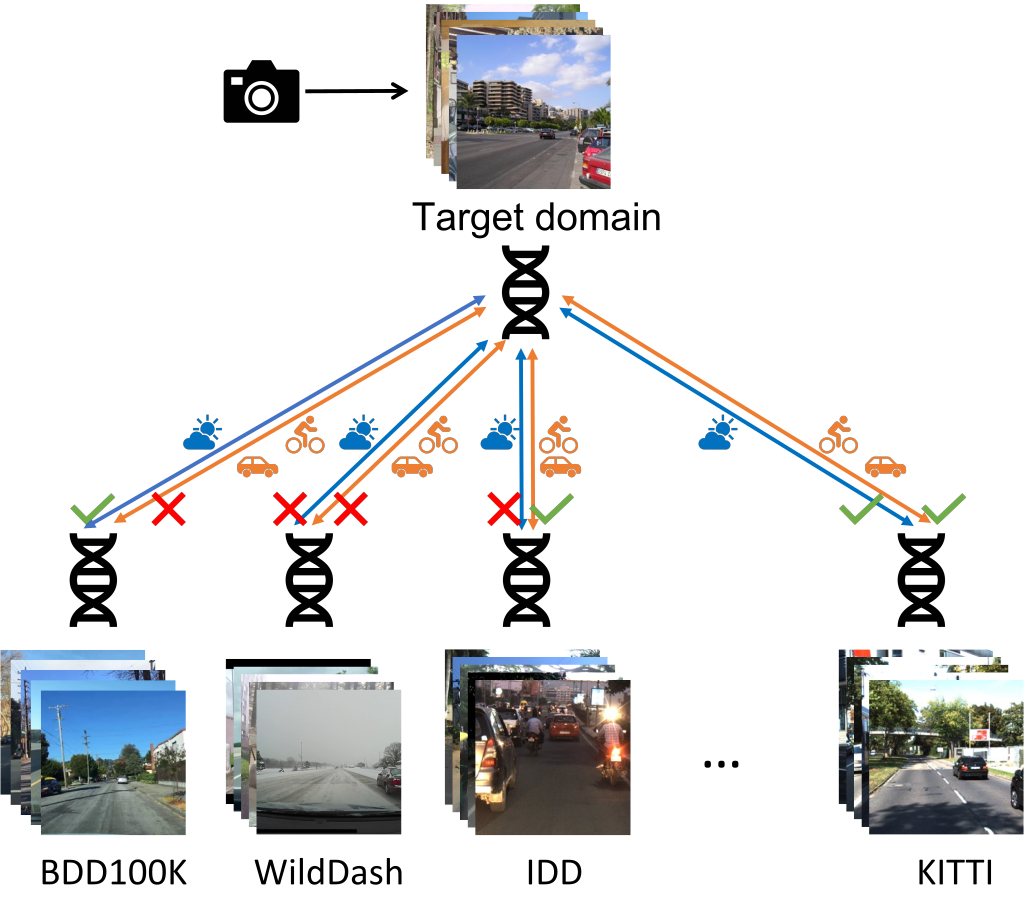
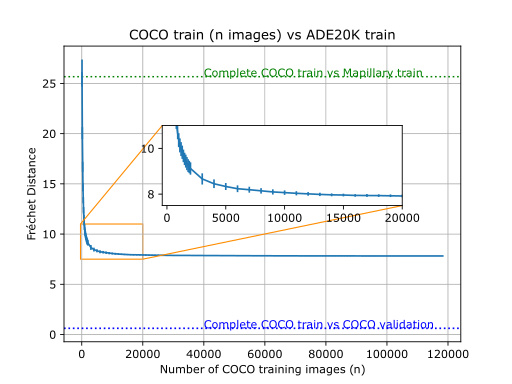
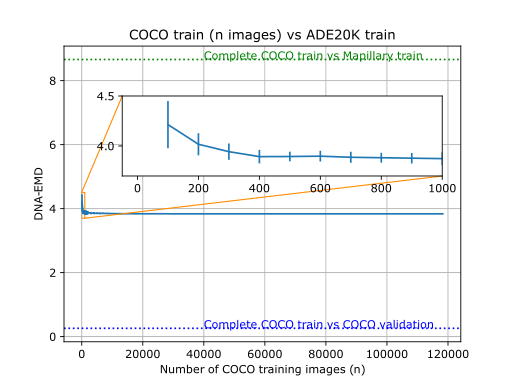
Example usage comparing different datasets
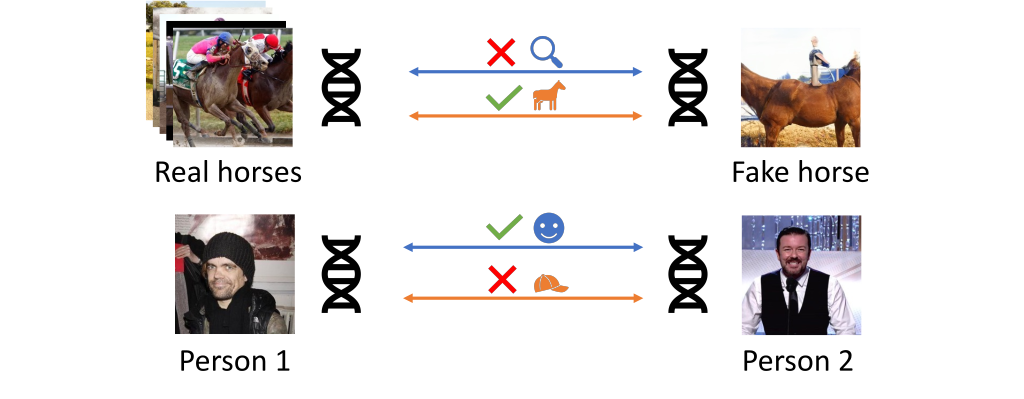
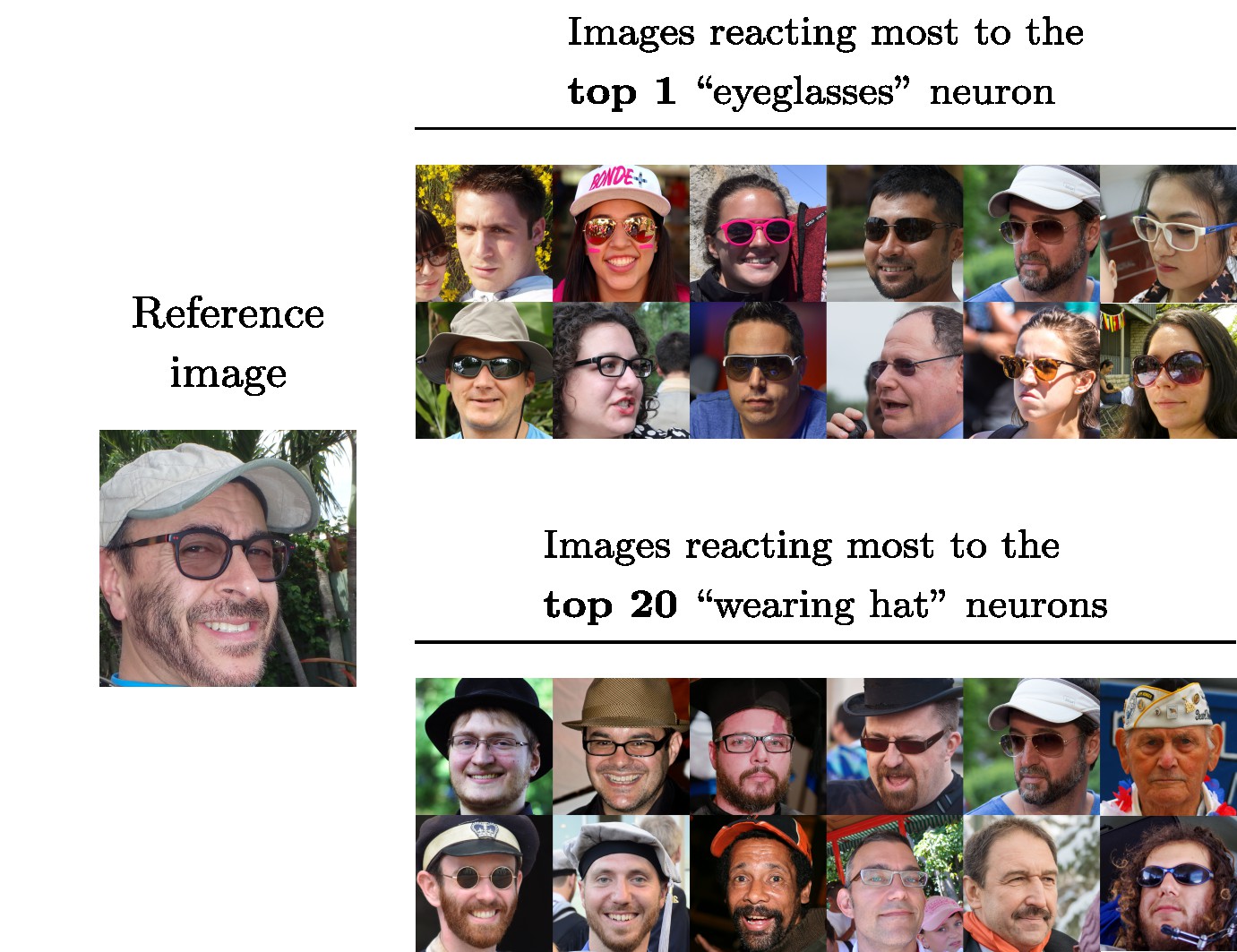
Example usage comparing an image to a dataset or pairs of images.
Visual DNAs allow you to generate compact granular representations of images and datasets. Visual DNAs can be compared conditionally thanks to the granularity they provide.
What is a Visual DNA?
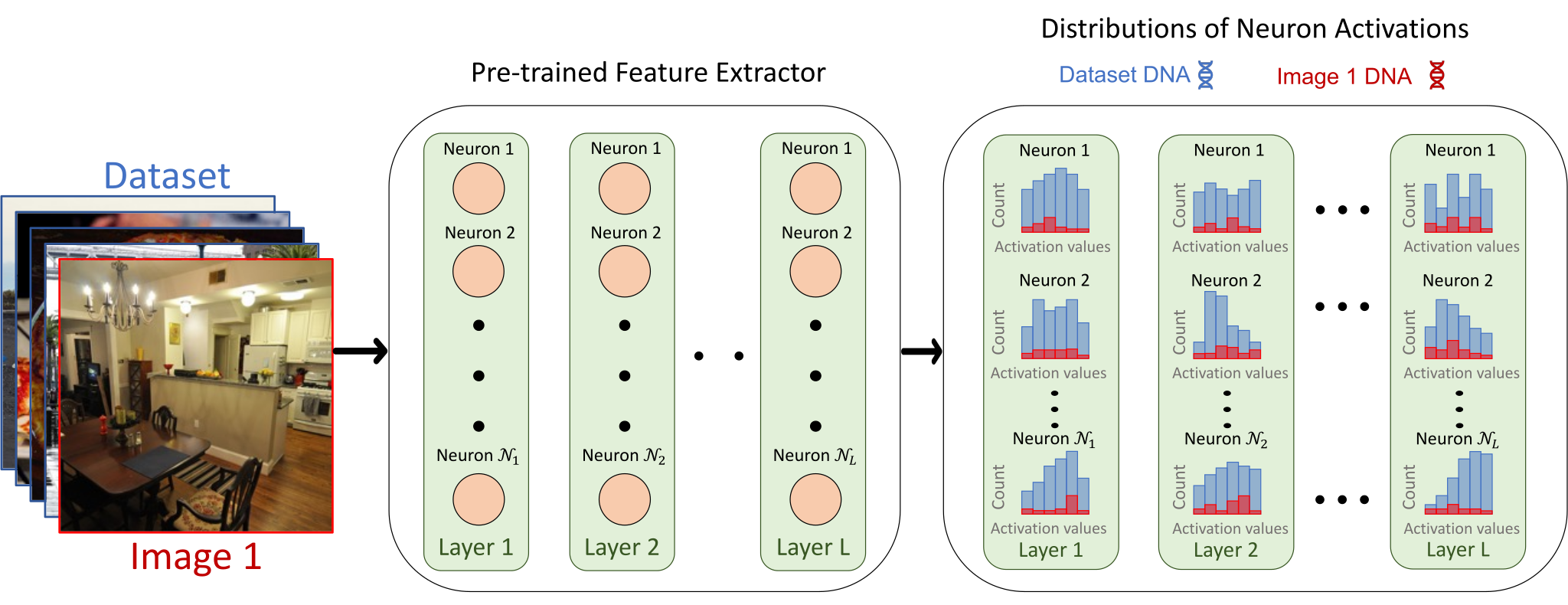
Visual DNAs consist of the Distributions of
Neuron Activations of a pre-trained frozen feature extractor when fed with
the images to represent. We then accumulate activations from different images using histograms or by fitting
a Gaussian.
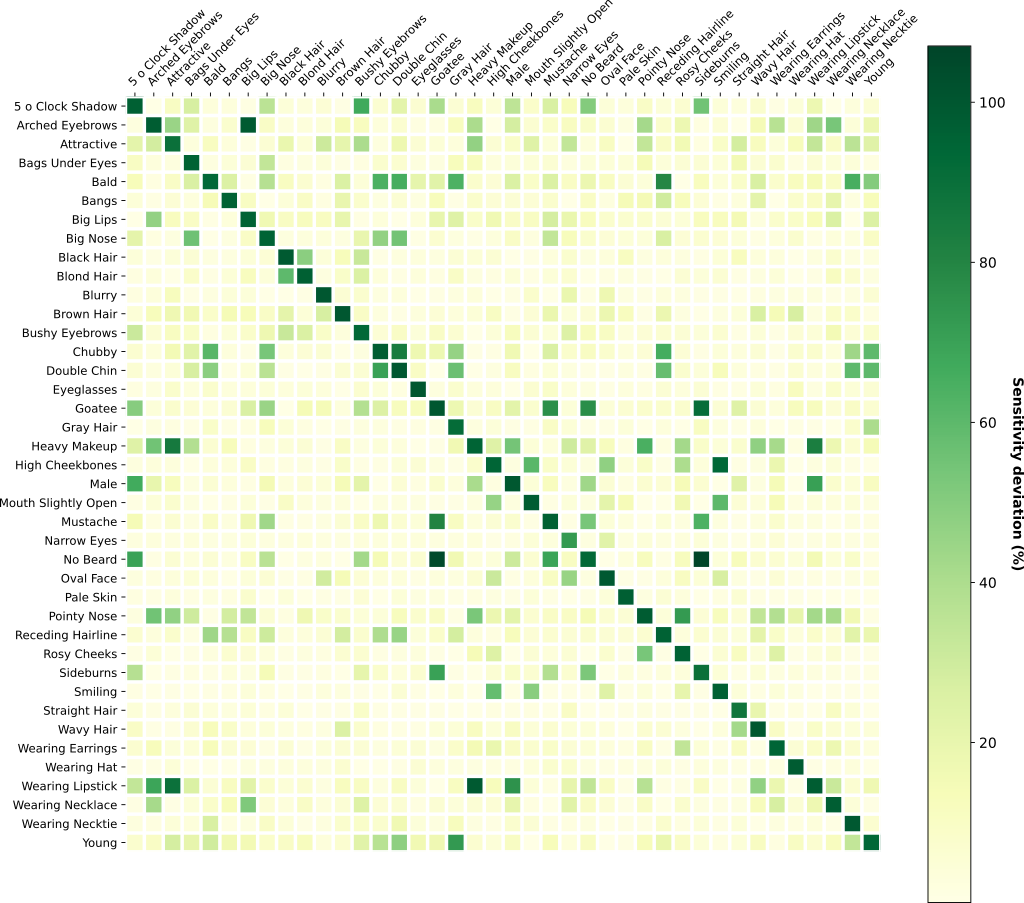
Comparing Visual DNAs comes down to comparing distributions (histograms or Gaussians) for each neuron. As
we don't expect all neurons of the pre-trained network to be sensitive to the particular properties we are
interested in comparing, this granularity allows to create custom comparisons that only rely on neurons of
interest.